Posts
My thoughts and ideas
Welcome to the blog
My thoughts and ideas
Introduction to bioinformatics for RNA sequence analysis
Description of the lab Welcome to the lab for Genome Visualization! This lab will introduce you to the Integrative Genomics Viewer, one of the most popular visualization tools for High Throughput Sequencing (HTS) data.
Lecture files that accompany this tutorial:
After this lab, you will be able to:
Things to know before you start:
The lab may take between 1-2 hours, depending on your familiarity with genome browsing. Do not worry if you do not complete the lab. It will remain available to review later.
There are a few thought-provoking Questions or Notes pertaining to sections of the lab. These are optional, and may take more time, but are meant to help you better understand the visualizations you are seeing. These questions will be denoted by boxes, as follows: Question(s):
Thought-provoking question goes here.`
Ability to run Java
Note that while most tutorials in this course are performed on the cloud, IGV will always be run on your local machine
Note a version of this tutorial can also be performed directly in your web browser at sandbox.bio IGV Intro.
This tutorial was most recently updated for IGV v2.16.2, which is available on the IGV Download page. It is recommended that you use this version. Most other recent versions will work but their may be slight differences.
We will be using publicly available Illumina sequence data from the HCC1143 cell line. The HCC1143 cell line was generated from a 52 year old caucasian woman with breast cancer. Additional information on this cell line can be found here: HCC1143 (tumor, TNM stage IIA, grade 3, primary ductal carcinoma) and HCC1143/BL (matched normal EBV transformed lymphoblast cell line).
We will be visualizing read alignments using IGV, a popular visualization tool for HTS data.
First, lets familiarize ourselves with it.
By default, IGV loads the Human GRCh38/hg38 reference genome. If you work with another version of the human genome, or another organism altogether, you can change the genome by clicking the drop down menu in the upper-left. For this lab, we will be using Human GRCh37/hg19.
We will also load additional tracks from Server using (File -> Load from Server...):
Load hg19 genome and additional data tracks
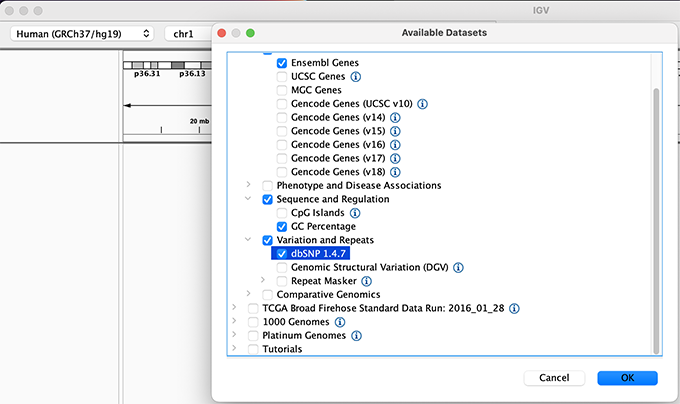
You should see listing of chromosomes in this reference genome. Choose 1, for chromosome 1.
Chromosome chooser

Navigate to chr1:10,000-11,000 by entering this into the location field (in the top-left corner of the interface) and clicking Go. This shows a window of chromosome 1 that is 1,000 base pairs wide and beginning at position 10,000.
Navigition using Location text field. Sequence displayed as thin coloured rectangles.
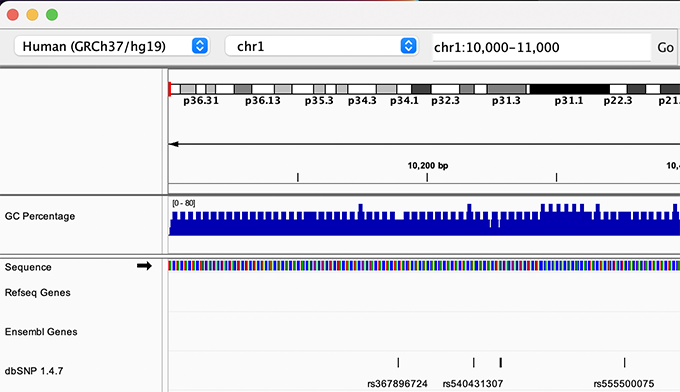
IGV displays the sequence of letters in a genome as a sequence of colours (e.g. A = green, C = blue, etc.). This makes repetitive sequences, like the ones found at the start of this region, easy to identify. Zoom in a bit more using the + button to see the individual bases of the reference genome sequence.
You can navigate to a gene of interest by typing it in the same box the genomic coordinates are in and pressing Enter/Return. Try it for your favourite gene, or BRCA1 if you can not decide.
Gene model

Genes are represented as lines and boxes. Lines represent intronic regions, and boxes represent exonic regions. The arrows indicate the direction/strand of transcription for the gene. When an exon box become narrower in height, this indicates a UTR.
When loaded, tracks are stacked on top of each other. You can identify which track is which by consulting the label to the left of each track.
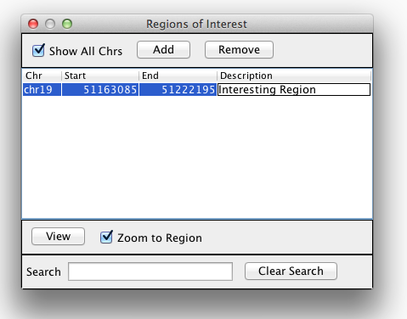
Sometimes, it is really useful to save where you are, or to load regions of interest. For this purpose, there is a Region Navigator in IGV. To access it, click Regions > Region Navigator. While you browse around the genome, you can save some bookmarks by pressing the Add button at any time.
Bookmarks in IGV
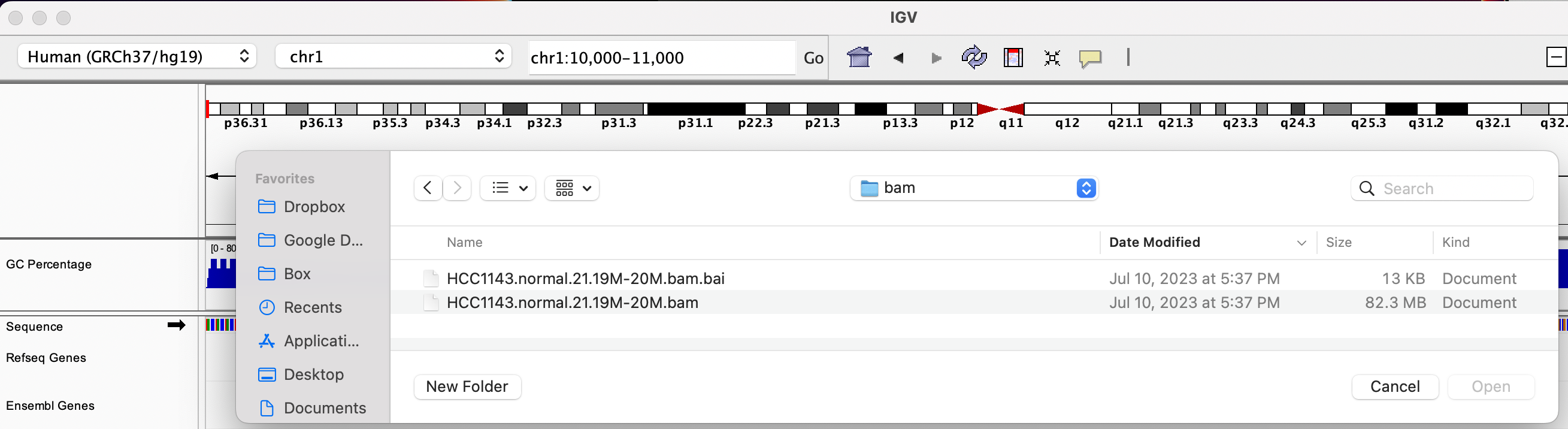
We will be using the breast cancer cell line HCC1143 to visualize alignments. For speed, only a small portion of chr21 will be loaded (19M:20M).
HCC1143 Alignments to hg19:
Copy the files to your local drive, and in IGV choose File > Load from File..., select the bam file, and click OK. Note that the bam and index files must be in the same directory for IGV to load these properly.
Load BAM track from File
Navigate to a narrow window on chromosome 21: chr21:19,480,041-19,480,386.
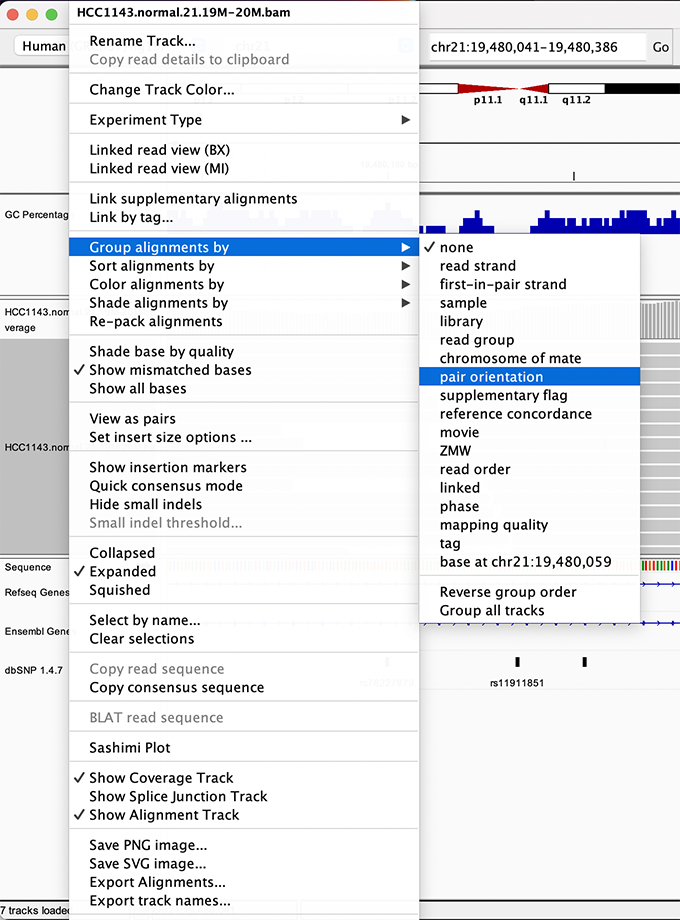
To start our exploration, right click on the read alignment track, and select the following options:
Sort alignments by -> start location
Group alignments by -> pair orientation
Experiment with the various settings by right clicking the read alignment track and toggling the options. Think about which would be best for specific tasks (e.g. quality control, SNP calling, CNV finding).
Changing how read alignments are sorted, grouped, and colored
You will see reads represented by grey or white bars stacked on top of each other, where they were aligned to the reference genome. The reads are pointed to indicate their orientation (i.e. the strand on which they are mapped). Mouse over any read and notice that a lot of information is available. To toggle read display from hover to click, select the yellow box and change the setting.
Changing how read information is shown (i.e. on hover, click, never)
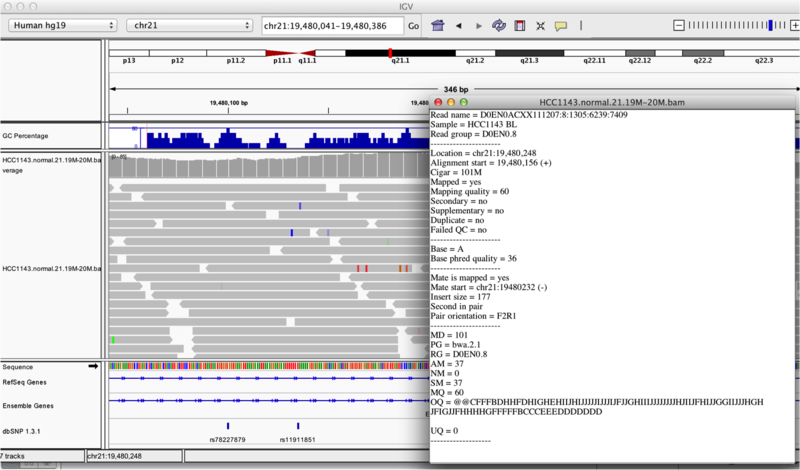
Once you select a read, you will learn what many of these metrics mean, and how to use them to assess the quality of your datasets. At each base that the read sequence mismatches the reference, the colour of the base represents the letter that exists in the read (using the same colour legend used for displaying the reference).
Viewing read information for a single aligned read
In this section we will be looking in detail at 8 positions in the genome, and determining whether they represent real events or artifacts.
Navigate to region chr21:19,479,237-19,479,814
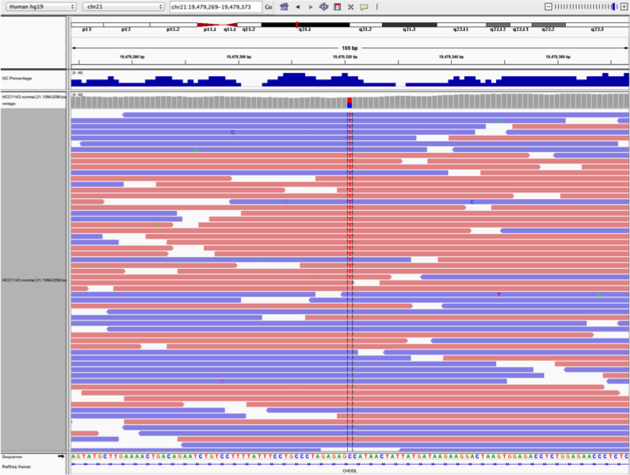
Note two heterozygous variants, one corresponds to a known dbSNP (G/T on the right) the other does not (C/T on the left)
Zoom in and center on the C/T SNV on the left, sort by base (window chr21:19,479,321 is the SNV position)
Sort alignments by base
Color alignments by read strand
Example1. Good quality SNVs/SNPs
Notes:
Question(s):
* What does *Shade base by quality* do? How might this be helpful?
* How does Color by *read strand* help?
Navigate to position chr21:19,518,412-19,518,497
Example 2a
read strand
A within the homopolymer run (chr21:19,518,470), and Sort alignments by -> base
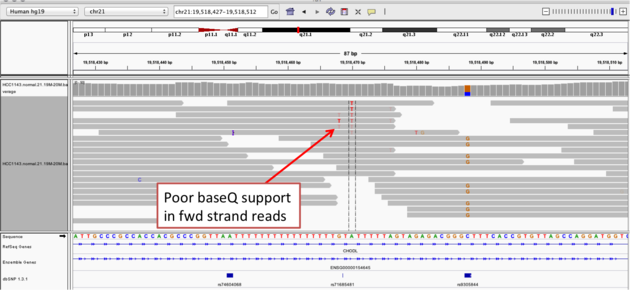
Example 2b
chr21:19,518,452), and Sort alignments by -> base
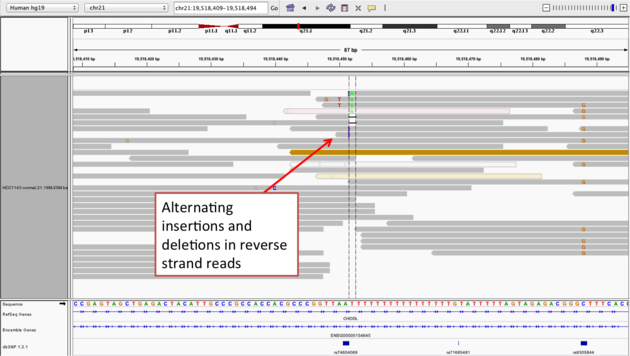
Notes:
Navigate to position chr21:19,611,925-19,631,555. Note that the range contains areas where coverage drops to zero in a few places.
Example 3
Collapsed viewColor alignments by -> insert size and pair orientation
Group alignments by -> none
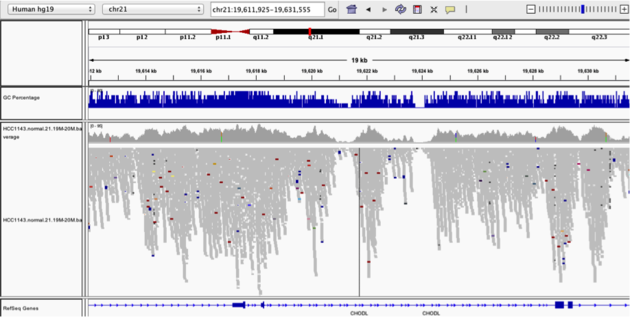
Question:
* Why are there blue and red reads throughout the alignments?
Navigate to region chr21:19,666,833-19,667,007
Example 4
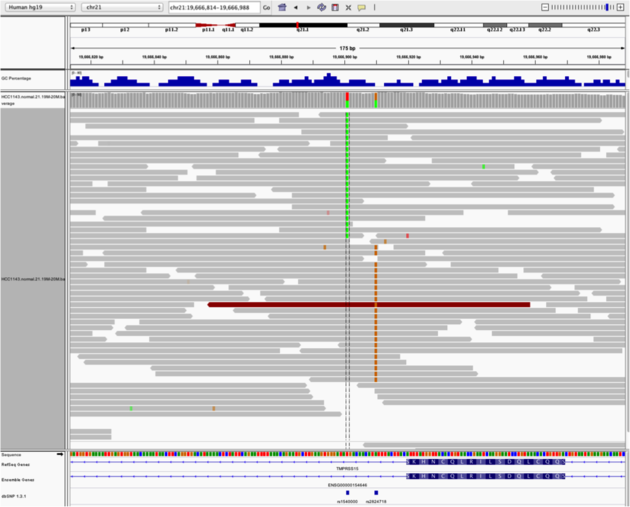
Note:
Navigate to region chr21:19,800,320-19,818,162
File -> Load from server...)Load repeats

Example 5
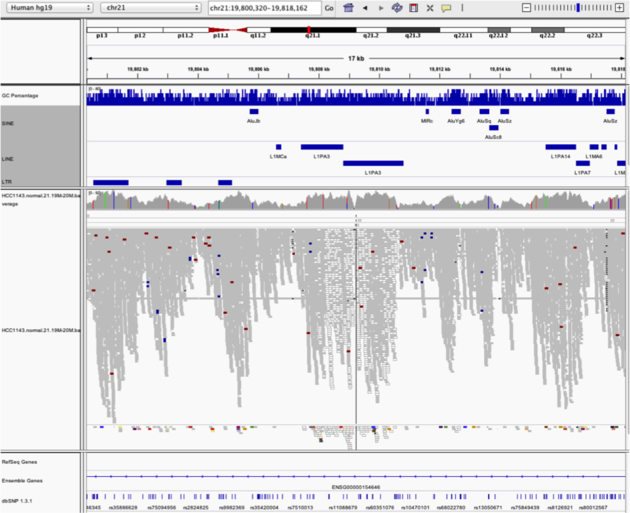
Notes:
Navigate to region chr21:19,324,469-19,331,468
Example 6
View as Pairs and Expanded viewColor alignments by -> insert size and pair orientation
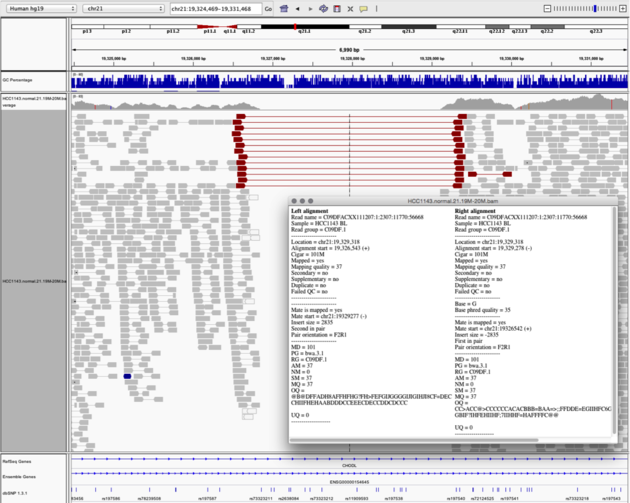
Notes:
Navigate to region chr21:19,102,154-19,103,108
Example 7

Notes:
Navigate to region chr21:19,089,694-19,095,362
Example 8
Group alignments by -> pair orientation
Color alignments by -> insert size and pair orientation
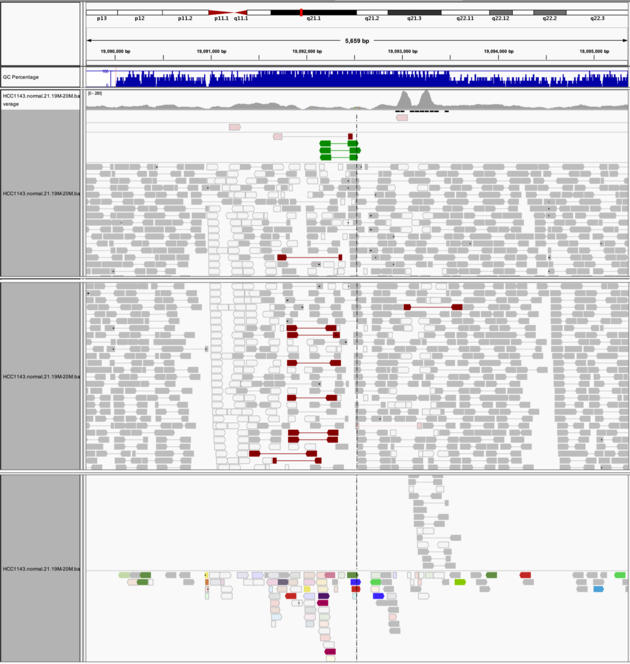
Notes:
We can use the Tools menu to invoke running a batch script. Batch scripts are described on the IGV website:
Download the batch script and the attribute file for our dataset:
Hint: You can use the curl -L -O with the above URLs to download the files using the terminal!
Now run the file from the Tools menu:
Automation
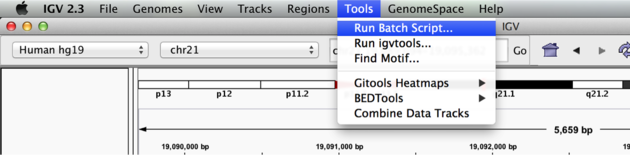
Notes:
Malachi Griffith, Sorana Morrissy, Jim Robinson, Ben Ainscough, Jason Walker, Obi Griffith, Kartik Singhal