RNA-seq
Welcome to the RNA-seq Bioinformatics Course. Use the course page to navigate your way through all exercises. Each page has a course link at the top to bring you back to the table of contents.
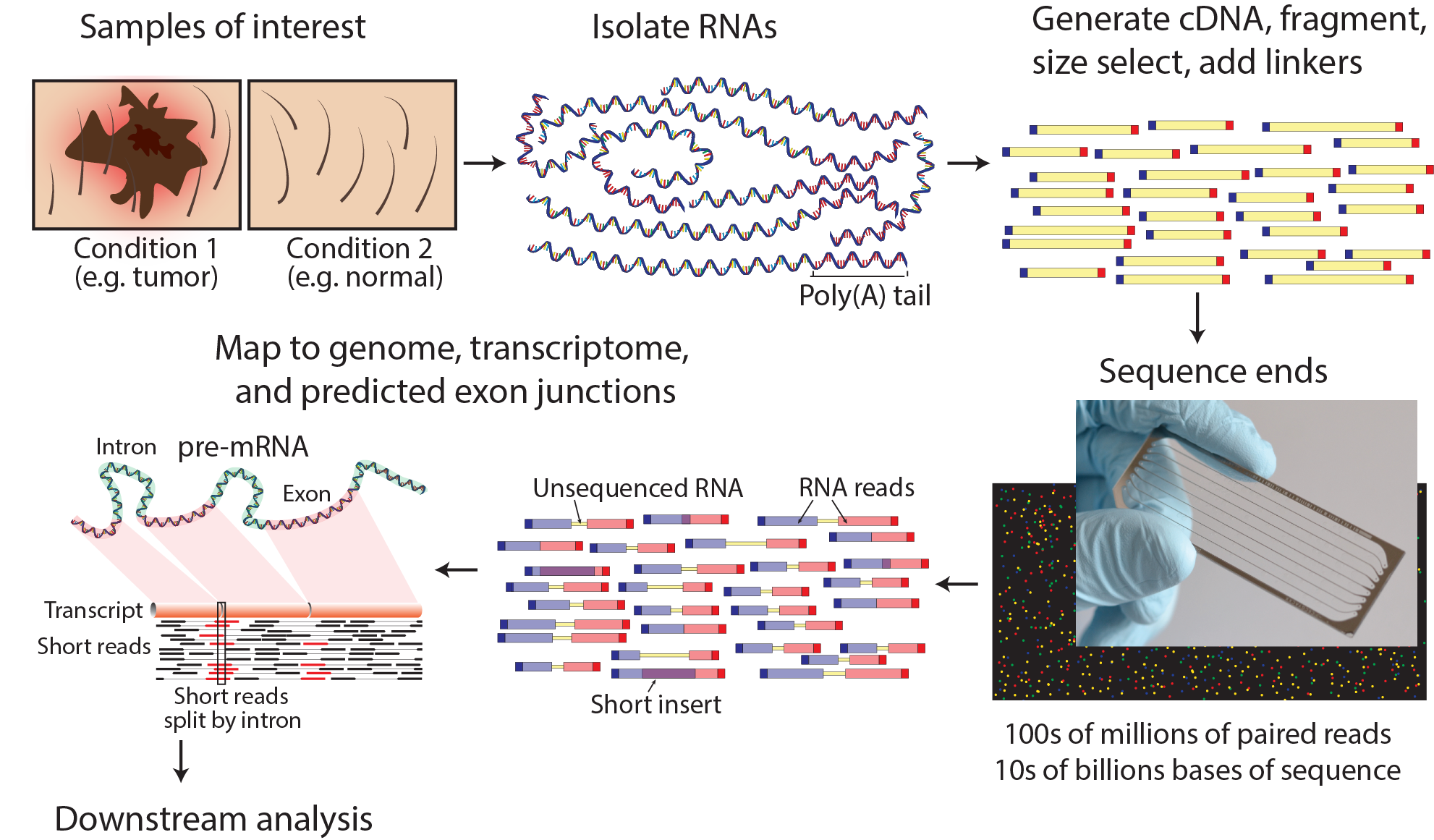
The age of affordable massively parallel sequencing has exponentially increased the availability of transcriptome profiling. RNA sequencing (RNA-seq) has rapidly become the assay of choice for robustly interrogating RNA transcript abundance and diversity. Similarly single cell RNA sequencing (scRNA-seq) has been widely adopted for experimental systems where it is crucial to understand a complex mixture of cell types. We have therefore developed this course to provide an introduction to RNA-seq and scRNA-seq data analysis concepts followed by integrated tutorials demonstrating the use of popular bioinformatics analysis packages. The tutorials are designed as self-contained units that include example data (Illumina paired-end RNA-seq data) and detailed instructions for installation of all required bioinformatics tools (HISAT, StringTie, Kallisto, etc.). Participants will gain practical experience and skills to be able to:
- Perform command-line Linux based analysis on the cloud
- Assess quality of RNA-seq data
- Align RNA-seq data to a reference genome
- Estimate known gene and transcript expression
- Perform differential expression analysis
- Perform reference guided and de novo assembly of transcript sequences
- Discover novel isoforms
- Perform scRNA data processing, cell type identification and differential expression analysis of cell clusters
- Visualize and summarize the output of RNA-seq and scRNA-seq analyses in R
A version of this workshop was accompanied by a publication. If you find the materials here useful, please cite the following: Malachi Griffith*, Jason R. Walker, Nicholas C. Spies, Benjamin J. Ainscough, Obi L. Griffith*. 2015. Informatics for RNA-seq: A web resource for analysis on the cloud. PLoS Comp Biol. 11(8):e1004393. *To whom correspondence should be addressed: E-mail: mgriffit[AT]genome.wustl.edu, ogriffit[AT]genome.wustl.edu
These materials have been developed with the support of AWS Cloud Credits for Research, the Canadian Bioinformatics Workshops, and Cold Spring Harbor Laboratory. See Acknowledgements for more details.
This workshop has been delivered many times and continues to evolve. A complete list of these past workshops is available here.
A legacy version of the online resource is also available here.
We are constantly improving the course material, if you notice a problem or have a suggestion for improvement please let us know by making a github issue at: https://github.com/griffithlab/rnabio.org/issues
